Training ivis on Out-of-memory Datasets
Introduction
Out-of-memory Datasets
Some datasets are so large that it becomes infeasible to load them into memory all at the same time. Other visualisation techniques might only be able to run on a smaller subset of the data; however, this runs the risk of potentially missing out on important smaller patterns in the data.
ivis was developed to address the issue of dimensionality reduction in very large datasets
through batch-wise training of the neural network on data stored HDF5 format.
Since training occurs in batches, the whole dataset does not need to
be loaded into memory at once, and can instead be loaded from disk in
chunks. In this example, we will show how ivis can scale up and
be used to visualize massive datasets that don’t fit into memory.
Example
Data Selection
In this example we will make use of the KDD Cup 1999 dataset. Although the dataset can be easily read-in to RAM, it provides a toy example for a general use case. The KDD99 dataset contains network traffic, with the competition task being to detect network intruders. The dataset is unbalanced, with the majority of traffic being normal.
Data Preparation
To train ivis on an out-of-memory dataset, the dataset must first be
converted into the h5 file format. There are numerous methods of doing
this using various external tools such as Apache Spark. In this example, we will assume that
the dataset has already been preprocessed and converted to .h5 format.
Dimensionality Reduction
To train on a h5 file that exists on disk you need the h5py package.
A HDF5 dataset stored inside a file can be directly passed to an Ivis object’s fit and transform methods.
We will train ivis in unsupervised mode for 5 epochs to speed up training;
other hyperparameters are left at their default values.
Note
When training on a h5 dataset, we recommend to use the shuffle_mode='batch' option in the fit method. This will speed up the training process by shuffling batches of data, rather than shuffling across the whole dataset.
import h5py
with h5py.File(h5_filepath, 'r') as f:
X = f['data']
y = f['labels']
model = Ivis(epochs=5)
model.fit(X, shuffle_mode='batch') # Shuffle batches when using h5 files
y_pred = model.transform(X)
Visualisations
plt.figure()
plt.scatter(x=y_pred[:, 0], y=y_pred[:, 1], c=y)
plt.set_xlabel('ivis 1')
plt.set_ylabel('ivis 2')
plt.show()
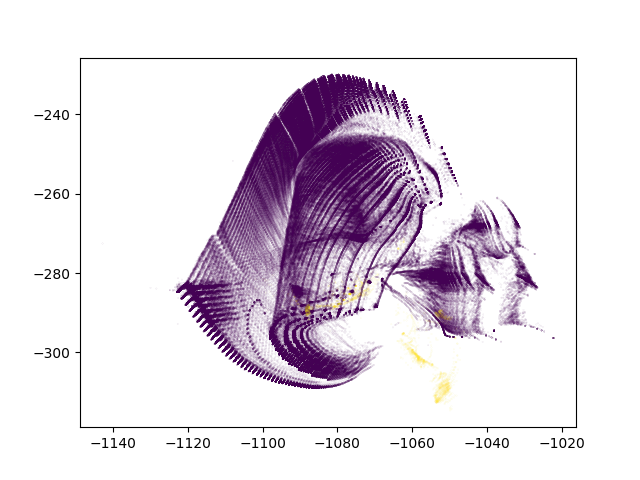
With anomalies being shown in yellow, we can see that ivis is able to pin point anomalous observations.
Conclusions
ivis
is able to scale and deal with the massive, out-of-memory datasets
found in the real world by training directly on h5 files. Additionally, it can effectively learn
embeddings in an unbalanced dataset without labels.
Advanced: Custom dataset loaders
In addition to h5 files, ivis is also able to train on arbitrary out-of-memory datasets using custom classes that implement the ivis.data.sequence.IndexableDataset class methods. For example, the ivis.data.sequence.ImageDataset inherits from the IndexableDataset and reads in images from disk when indexed, allowing for training on image datasets of any size. The instance of ImageDataset is simply provided to ivis as the argument to the fit or transform methods.
By writing a custom class tailored to your specific scenario you can use ivis with whatever data storage you are using. For example, it’s possible to have ivis train directly on data stored within a database with just a few lines of code.